Imaging Software Bring the Brain into Fuller Focus
Innovative software developed at the Oxford Centre for Functional MRI of the Brain (FMRIB) is on track to improve our understanding of the mind’s innermost wirings – opening up exciting possibilities in neuroscience.
Funded by the University of Oxford and the Human Connectome Project (see below), a team including research student Moisés Hernández Fernández, research fellow Dr Stamatios Sotiropoulos, Prof. Stephen Smith and Prof. Michael Giles has developed parallel computation algorithms that vastly accelerate the processing of Data Mountains generated by a key medical imaging technology.
This breakthrough, in which the Emerald supercomputer funded by the Engineering and Physical Sciences Research Council (EPSRC) has played a vital role, will enable exploration of brain structure in unprecedented detail and potentially provide a practical tool for clinical use.
New Frontiers of Neuroscience
Developing a deeper understanding of how the brain works is arguably one of the biggest scientific challenges of the 21st century. Huge strides have been made but much remains to be discovered in terms of how information is transferred both within the brain and with other parts of the body. Answering these questions will not just enhance our conception of what it actually means to be human – it is also critical to generating fresh insights into neurological disorders such as Alzheimer’s disease and multiple sclerosis. In this context:
- Cutting-edge imaging and modelling techniques can make a key
contribution by enabling non-invasive exploration of living brains, helping to unravel the microstructures and the unimaginably complex ‘wiring’ that underpin the brain and its workings. - Diffusion magnetic resonance imaging (dMRI) – including a technique known as white matter tractography – is a proven means of generating 3D representations of pathways in the brain’s white matter that mediate information transfer between remote brain regions.
“Our aim has been to devise software that takes a different approach, enabling dMRI brain data to be analysed very quickly and very accurately and taking the contribution of computational science to clinical neuroscience to the next level” – Moises Hernandez Fernandez, research student
A major problem, however, has been difficulty in handling the vast datasets produced by dMRI when scanning a highly sophisticated organ like the brain. The sheer length of time it takes to process and analyse this data has restricted the use of dMRI in this application as well as the level of detail that can be extracted, both in academic circles and in clinical environments.
“Conventional Central Processing Units, or CPUs, that are the mainstay of computers have more or less reached their limits,” Moisés Hernández Fernández explains. “This meant that the only solution in terms of continuing improvement of computing performance was to increase the number of computing units, leading to the emergence of multi-core CPUs and parallel computing architectures.Our aim here at FMRIB has been to devise software that takes a different approach, enabling dMRI brain data to be analysed very quickly and very accurately and taking the contribution of computational science to clinical neuroscience to the next level.”
Map showing the estimated principal diffusion orientations in each voxel of the brain: medial-lateral orientations are coloured red, superior-inferior orientations are blue and anterior-posterior orientations are green. Producing such a map is very expensive computationally, making it beneficial to use GPUs which are 150 times quicker than CPUs.
Close-up of the yellow box in the previous image, showing details of the estimated principal diffusion orientations in a region within the brain’s Centrum Semiovale.
A Parallel Path to Better Imaging
The team’s solution has involved rebuilding dMRI data analysis algorithms so that they can run on Graphics Processing Units (GPUs). Containing thousands of computer cores, GPUs are game-changing electronic circuits first developed over a decade ago and specifically designed to handle multiple tasks simultaneously. The parallel architectures they provide can, depending on the algorithm used, process data much faster than CPUs.
Development and testing of the software involved several months’ utilisation of over 100 GPUs at the Emerald supercomputer developed by Bristol, Oxford and Southampton Universities and University College London and located at Rutherford Appleton Laboratory in South Oxfordshire. The team subsequently invested in a number of GPUs for their own laboratory at FMRIB, which is based at Oxford’s John Radcliffe Hospital.
“Parallel computing architectures make it possible to process massive volumes of data on practical timescales”
Dr. Stamatios Sotiropoulos, Research Fellow
“Parallel computing architectures make it possible to process massive volumes of data on practical timescales,” says Stamatios Sotiropoulos. “Our algorithm designs enable GPUs to work around 150 times faster than CPU implementations. Such accelerations change the perspective of what is computationally feasible. This is not only beneficial for sheer data processing using existing paradigms, but also affects the amount of exploration that can be performed when researching new methods and technology.”
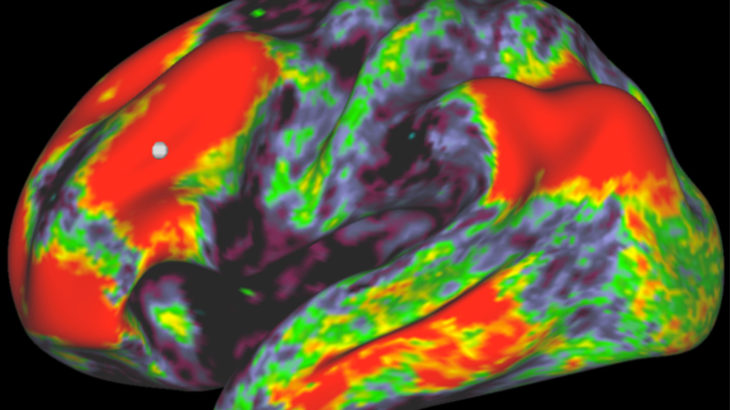
Figure 3: Connectivity matrix of the brain, with red representing high confidence in a connection and blue representing low confidence. Calculating this matrix is blalnk spacevery expensive computationally, but using GPUs can accelerate the process by a factor of 50. Figure 4: Representation of a row within the matrix in the previous image. The different colours represent the degree of confidence in a connection.
Building a Clearer Picture
Thanks to the speed with which it can extract important information and insights about brain wiring, the software is being harnessed by pioneering initiatives including:
- The Human Connectome Project – a 5-year study aiming to produce a comprehensive, ground-breaking ‘brain map’ depicting neural connections in the brain and the pathways that underlie brain function and behaviour.
- The Developing Human Connectome Project – focused on creating a dynamic map of neural connections in human foetuses and neonates between 20 and 44 weeks after conception, providing insight on developmental processes and associated pathologies.
- The UK Biobank Project – developing a huge resource of health-related data gathered from 100,000 volunteers to help explain why some people develop particular diseases while others do not.
“We’ve already successfully tested our software on massive high-resolution datasets generated by the Human Connectome Project and demonstrated that it can complete in a matter of hours tasks that would otherwise take weeks,” Hernández Fernández concludes. “Once we’ve finished optimising the software, we’re fully confident that we’ll start to see it deliver real dividends in a very wide range of applications.”
- Hernández, G. D. Guerrero, J. M. Cecilia, J. M. Garcia, A. Inuggi, S. Jbabdi, T. E. J. Behrens and S. N. Sotiropoulos. Accelerating Fibre Orientation Estimation from Diffusion Weighted Magnetic Resonance Imaging Using GPUs. PLOS ONE, 29 April 2013; DOI: 10.1371/journal.pone.0061892
- N. Sotiropoulos, S. Jbabdi, J. Xu, J.L. Andersson, S. Moeller, E.J. Auerbach, M.F. Glasser, S.M. Hernandez, G. Sapiro, M. Jenkinson, D.A. Feinberg, E. Yacoub, C. Lenglet, D.C. Van Essen, K. Ugurbil and T.E. Behrens. Advances in Diffusion MRI Acquisition and Processing in the Human Connectome Project.NeuroImage, 2013, DOI: 10.1016/j.neuroimage.2013.05.057
Contacts

Moisés Hernández Fernández
Oxford Centre for Functional MRI of the Brain (FMRIB), John Radcliffe Hospital
Dr Stamatios Sotiropoulos
Oxford Centre for Functional MRI of the Brain (FMRIB), John Radcliffe Hospital

