How HPC is Boosting Cutting-Edge Biology
High Performance Computing (HPC) is adding momentum to the evolution of biological science and a revolution in our understanding of how biological systems function – as pioneering research at UCL is clearly demonstrating.
To drive their work forward, Dr Chris Barnes and his Computational Systems and Synthetic Biology Group make extensive use of large HPC systems, especially Emerald, the SES GPU-accelerated supercomputer, established using funding from the Engineering and Physical Sciences Research Council (EPSRC) .
The group is at the leading edge of the fast-developing field of quantitative biology, a discipline that uses measurable data and mathematical modelling to reveal the secrets of the living world – with Emerald’s sheer computational power and speed helping to generate insights that could lead to dramatic advances in spheres such as drug development and gene therapy.
Biology Breaks Out
Biology is one of the great natural sciences. Traditionally, its main focus has been on identifying and characterising the different elements within biological systems. But how do these elements actually combine and work together? This key gap in understanding helped to trigger the comparatively recent emergence of quantitative biology, which strives to:
- Establish firm mathematical and physical foundations enabling biological science to secure new insights and break into new areas.
- Harness a comprehensive array of computational, mathematical and statistical tools and techniques to make this possible.
“My group supplements experimental methods with mathematical modelling and statistical analysis,” Chris Barnes explains. “We then apply these tools to biological systems and synthetic biology – an emerging discipline specialising in the design and construction of artificial biological systems and the redesign of existing ones, with potential uses not just in medicine and healthcare but also in areas such as energy and the environment.”
A key element in this work, which is underpinned by UK Research Council funding and support from the Wellcome Trust, involves building mathematical models of biological systems to show how they function and might be improved. “We use models to make predictions that we then test through physical experiments, which in turn generate results that we feed back into the models,” says Dr Barnes. “It’s a process where modelling and experimentation are continually reinforcing one another.”
“Some of our projects simply couldn’t and wouldn’t have happened without the computing power that Emerald provides”
Dr Chris Barnes, Lecturer in Systems Biology
Emerald Shines Vital Light on Key Questions
In a field where computational tools naturally play a pivotal role, HPC has delivered a step change in capabilities enabling the team to carry out increasingly complex and sophisticated modelling with phenomenal speed and accuracy. In this context, Emerald, located at Rutherford Appleton Laboratory in Oxfordshire and the UK’s most powerful supercomputer based on Graphics Processing Units (GPUs), has been a game-changer.
“We first harnessed Emerald in summer 2012 as one of its very first pilot users,” Chris Barnes confirms. “Since then, Emerald has become absolutely integral to the challenging work we do using complicated non-linear models to investigate different types of biological processes and to improve understanding of their complexities.”
As the following recent examples show, this cutting-edge 84-node cluster is adding real value to the ground-breaking research being pursued by the Barnes group:
- DNA repair and mutation:
The DNA strands in living cells are crucial carriers of genetic information, but they can be damaged by a range of sources originating both inside and outside the cell. Complex repair systems have evolved over millions of years, varying according to factors such as the nature of the damage and the stage in the cell’s life cycle at which it occurs. In some cases, though, the repair process can misfire and result in mutations – some of which, like cancer, can have serious health implications.“We’ve recently used Emerald to help us apply modelling and statistical methods to genomic and experimental data, to shed light on different DNA repair and mutation mechanisms,” says Dr Mae Woods, an applied mathematician in the group. “For example, Chris Barnes and I have made extensive use of the facility to develop a computational model of DNA repair.” - Engineering bacteria:
The engineering of so-called ‘commensal’ bacteria (which can exist inside other organisms without harming them) has exciting potential in terms of creating new forms of therapy and revolutionary tools for disease diagnosis. Such bacteria, however, need to be capable of functioning in wide-ranging and ever-changing temperature, pH and other conditions – which makes it vital to be able to predict which designs of bacteria will be the most robust.
“With Emerald’s help, we’ve developed and tested design methods that allow the selection of the genetic networks incorporated into engineered bacteria to be based on the networks’ robustness to external noise”, says PhD student Miriam Leon.
Biology Benefits
“We’ve used Emerald to help us apply modelling and statistical methods to genomic and experimental data, to shed light on different DNA repair and mutation mechanisms”
Dr Mae Woods, Research Associate in Mathematical Modelling
By adding an extra component to the group’s toolkit, Emerald is certainly meeting its goal of delivering super-fast processing that accelerates research and solves problems quicker. “Some of our projects simply couldn’t and wouldn’t have happened without the computing power that Emerald provides,” Chris Barnes affirms unequivocally.
“Take our recent research into transcriptional oscillators as part of our work on bacteria. Some of the simulations that we completed in a few hours using Emerald would have taken up to a week running on single GPU cards, and generating the results required hundreds of runs. But just as important as raw computing power is the fact that Emerald is so accessible, easy to use and underpinned by such excellent technical support – all of which makes a massive difference to our ability to meet the challenges presented by this fascinating and increasingly influential field of biology.”
Discover More
- Woods, M. Leon, R. Perez-Carrasco and C.P. Barnes (2016). A Statistical Approach Reveals Designs for the Most Robust Stochastic Gene Oscillators.
ACS Synthetic Biology (in press), bioRxiv. doi:http://dx.doi.org/10.1101/025056
- Woods and C.P. Barnes. Alternative End Joining Mechanisms Exhibit Varying Dynamics in Double Strand Break Repair. bioRxiv doi: http://dx.doi.org/10.1101/026070
- Cohen, A. Kicheva, A. Ribeiro, R. Blassberg, K.M. Page, C.P. Barnes and J. Briscoe (2015). Ptch1 and Gli Regulate Shh Signalling Dynamics via Multiple Mechanisms.
Nature Communications, 6. doi:10.1038/ncomms7709 - Cohen, K.M. Page, R. Perez-Carrasco, C.P. Barnes and J. Briscoe (2014).
A Theoretical Framework for the Regulation of Shh Morphogen-controlled Gene Expression. Development, 141(20), 3868-3878. doi:10.1242/dev.112573 - L. Woods, C. Carmona-Fontaine, C.P. Barnes, I.D. Couzin, R. Mayor and K.M. Page (2014). Directional Collective Cell Migration Emerges as a Property of Cell Interactions.
PLoS One, 9(9), e104969. doi: 10.1371/journal.pone.0104969
UCL Computational Systems and Synthetic Biology Group
Miriam LeonImage 1:
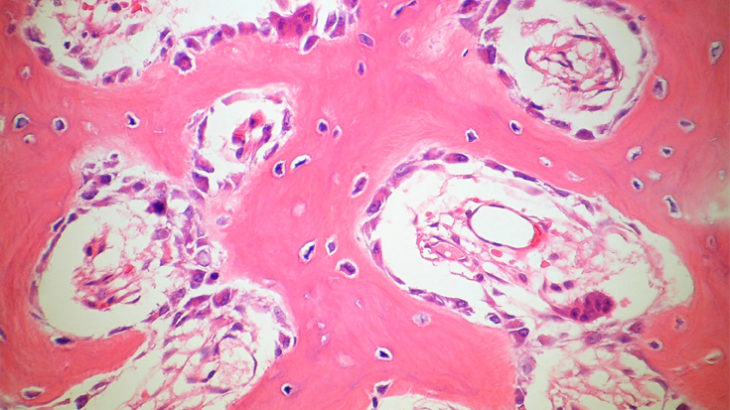
DNA repair in osteoblastoma (a rare type of benign bone tumour): cells are exposed to the DNA-damaging agent neocarzinostatin.
Image 2:
Example of a posterior distribution (a term used in the field of statistics) produced by ‘abc-smc’ analysis on Emerald.
Image 3:
Visual time series of a synthetic oscillator expressing green fluorescent protein in individual E. coli cells – the white arrow indicates the same single cell throughout the recording.
Image 4:
Exploring robustness – the system on the left oscillates in a smaller region of parameter space than the system on the right.
Contact

Dr Chris Barnes
Department of Cell & Developmental Biology, UCL

