Modelling mutations for cancer cure
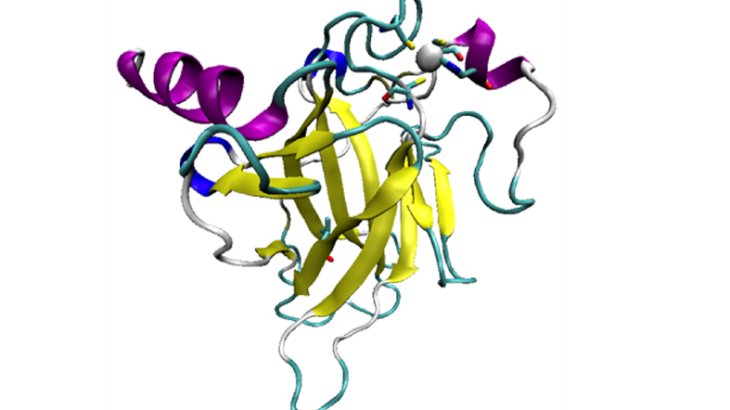
The p53 tumour suppressor protein, when it works properly, helps to prevent cancer. It does so by inducing the arrest of the cell cycle, by repairing DNA, and by causing apoptosis – a process of programmed cell death. However, p53 also regularly mutates, and either loses the ability to bind DNA, or is destabilised and unfolds, which induces the growth of DNA-damaged cells.
Half of all cancers are due to this mutation – so Zohra Ouaray, a PhD Research Student in the School of Chemistry at the University of Southampton, is using Emerald to model what happens and look at potential ways to prevent this happening.
“I’m basically trying to figure out how these mutations induce destabilisation of the structural behaviour of p53 wild-type. I’m using Emerald for the modelling because I need to do such long simulations, from 500 nanosecond to microsecond simulations, due to the slow motions that I’m studying”
Zahra Ouaray
Mutations inducing p53 destabilisations – called “structural mutants” – are investigated using these simulations. One of the first to be studied is known as V143A, a temperature-sensitive mutation. This induces destabilisation by creating a cavity in the protein at physiological temperatures, and also influences DNA-binding at lower temperatures.
95% of mutations occur in what is called the DNA-binding domain, of p53. Structural models of the p53 DBD, wild-type and mutant, were therefore chosen from the protein data bank
to be used in a Molecular Dynamics study. Different methods were tested, to propose a model as close as possible to the original protein. A range of simulations were then performed on these models, to create sample conformations at different temperatures and ion concentrations, and several analysis methods were used to study the stability of these samples.
“We want to be able to identify the unfolding pathway of the protein, to be able to potentially identify how small molecules or peptides can be used to restabilise some of the mutants”, Ouray says. In the long term, this could lead to novel, broadly applicable cancer treatments.
Now in the fourth year of her project, Ouaray says she has used Emerald since she began, including during two years when she was based in Singapore, collaborating with researchers at the Singapore A*STAR Bioinformatics Institute.
“The Emerald team helped me to use the software and have always helped with any problems I’ve had. Though it’s quite straightforward and I haven’t really had any issues – even when I was in Singapore, they made things easy for me to just connect from there” she says.
Visual representation of p53 DNA Binding Domain
She adds that “the Singapore team had its own supercomputer, of course, but it was faster to use Emerald – there were a lot of people using their computer. I didn’t want to have to wait – or to take space from the people there. The Emerald team have always been fast to answer any questions and fix any problems – and there have been very few problems, anyway. I couldn’t do my work without it, and they make it very easy.”
The authors would like to acknowledge that the work presented here made use of the Emerald High Performance Computing facility provided via the Science & Engineering South Consortium in partnership with STFC Rutherford Appleton Laboratory.

